Replication of Chapter 5 of Quantitative Social Science: An Introduction
Stefan Müller and Kenneth Benoit
Source:vignettes/pkgdown/replication/qss.Rmd
qss.RmdIn this vignette we show how the quanteda package can be used to replicate the text analysis part (Chapter 5.1) from Kosuke Imai’s book Quantitative Social Science: An Introduction (Princeton: Princeton University Press, 2017).
Download the Corpus
To get the textual data, you need to install and load the qss package first that comes with the book.
remotes::install_github("kosukeimai/qss-package", build_vignettes = TRUE)Section 5.1.1: The Disputed Authorship of ‘The Federalist Papers’
First, we use the readtext package to import the Federalist Papers as a data frame and create a quanteda corpus.
# use readtext package to import all documents as a dataframe
corpus_texts <- readtext::readtext(system.file("extdata/federalist/", package = "qss"))
# create docvar with number of paper
corpus_texts$paper_number <- paste("No.", seq_len(nrow(corpus_texts)), sep = " ")
# transform to a quanteda corpus object
corpus_raw <- corpus(corpus_texts, text_field = "text", docid_field = "paper_number")
# create docvar with authorship (used in Section 5.1.4)
docvars(corpus_raw, "paper_numeric") <- seq_len(ndoc(corpus_raw))
# create docvar with authorship (used in Section 5.1.4)
docvars(corpus_raw, "author") <- factor(NA, levels = c("madison", "hamilton"))
docvars(corpus_raw, "author")[c(1, 6:9, 11:13, 15:17, 21:36, 59:61, 65:85)] <- "hamilton"
docvars(corpus_raw, "author")[c(10, 14, 37:48, 58)] <- "madison"## AMONG the numerous advantages promised by a well-constructed Union, none
## deserves to be more accurately developed than its tendency to break and
## control the violence of faction. The friend of popular governments never
## Section 5.1.2: Document-Term Matrix
Next, we transform the corpus to a document-feature matrix.
dfm_prep (used in sections 5.1.4 and 5.1.5) is a dfm in
which numbers and punctuation have been removed, and in which terms have
been converted to lowercase. In dfm_papers, the words have
also been stemmed and a standard set of stopwords removed.
# transform corpus to a document-feature matrix
dfm_prep <- tokens(corpus_raw, remove_numbers = TRUE, remove_punct = TRUE) |>
dfm(tolower = TRUE)
# remove stop words and stem words
dfm_papers <- dfm_prep |>
dfm_remove(stopwords("en")) |>
dfm_wordstem("en")
# inspect
dfm_papers## Document-feature matrix of: 85 documents, 4,860 features (89.11% sparse) and 3 docvars.
## features
## docs unequivoc experi ineffici subsist feder govern call upon deliber new
## No. 1 1 1 1 1 1 9 1 6 3 5
## No. 2 0 3 0 0 2 9 1 1 1 2
## No. 3 0 1 0 0 2 20 0 0 0 1
## No. 4 0 2 0 0 0 21 1 0 0 0
## No. 5 0 1 0 0 0 3 0 0 0 0
## No. 6 0 3 0 0 0 4 0 4 0 1
## [ reached max_ndoc ... 79 more documents, reached max_nfeat ... 4,850 more features ]
# sort into alphabetical order of features, to match book example
dfm_papers <- dfm_papers[, order(featnames(dfm_papers))]
# inspect some documents in the dfm
head(dfm_papers, nf = 8)## Warning: nf argument is not used.## Document-feature matrix of: 6 documents, 4,860 features (90.51% sparse) and 3 docvars.
## features
## docs ` 1st 2d 3d 4th 5th abandon abat abb abet
## No. 1 0 0 0 0 0 0 0 0 0 0
## No. 2 0 0 0 0 0 0 0 0 0 0
## No. 3 0 0 0 0 0 0 0 0 0 0
## No. 4 0 0 0 0 0 0 0 0 0 0
## No. 5 0 1 0 0 0 0 0 0 0 0
## No. 6 0 0 0 0 0 0 0 0 0 0
## [ reached max_nfeat ... 4,850 more features ]The tm package considers features such as “1st” to be numbers, whereas quanteda does not. We can remove these easily using a wildcard removal:
dfm_papers <- dfm_remove(dfm_papers, "[0-9]", valuetype = "regex", verbose = TRUE)## dfm_remove() changed from 4,860 features (85 documents) to 4,855 features (85 documents)
head(dfm_papers, nf = 8)## Warning: nf argument is not used.## Document-feature matrix of: 6 documents, 4,855 features (90.50% sparse) and 3 docvars.
## features
## docs ` abandon abat abb abet abhorr abil abject abl ablest
## No. 1 0 0 0 0 0 0 0 0 1 0
## No. 2 0 0 0 0 0 0 1 0 0 0
## No. 3 0 0 0 0 0 0 0 0 2 0
## No. 4 0 0 0 0 0 0 0 0 1 1
## No. 5 0 0 0 0 0 0 0 0 0 0
## No. 6 0 0 0 0 0 0 0 0 0 0
## [ reached max_nfeat ... 4,845 more features ]Section 5.1.3: Topic Discovery
We can use the textplot_wordcloud() function to plot
word clouds of the most frequent words in Papers 12 and 24.
set.seed(100)
library("quanteda.textplots")
textplot_wordcloud(dfm_papers[c("No. 12", "No. 24"), ],
max.words = 50, comparison = TRUE)
Since quanteda cannot do stem completion, we will skip that part.
Next, we identify clusters of similar essay based on term frequency-inverse document frequency (tf-idf) and apply the -means algorithm to the weighted dfm.
# tf-idf calculation
dfm_papers_tfidf <- dfm_tfidf(dfm_papers, base = 2)
# 10 most important words for Paper No. 12
topfeatures(dfm_papers_tfidf[12, ], n = 10)## revenu contraband patrol excis coast trade per
## 19.42088 19.22817 19.22817 19.12214 16.22817 15.01500 14.47329
## tax cent gallon
## 13.20080 12.81878 12.81878
# 10 most important words for Paper No. 24
topfeatures(dfm_papers_tfidf[24, ], n = 10)## garrison dock-yard settlement spain armi frontier arsenal
## 24.524777 19.228173 16.228173 13.637564 12.770999 12.262389 10.818782
## western post nearer
## 10.806108 10.228173 9.648857We can match the clustering as follows:
k <- 4 # number of clusters
# subset The Federalist papers written by Hamilton
dfm_papers_tfidf_hamilton <- dfm_subset(dfm_papers_tfidf, author == "hamilton")
# run k-means
km_out <- stats::kmeans(dfm_papers_tfidf_hamilton, centers = k)
km_out$iter # check the convergence; number of iterations may vary## [1] 2
colnames(km_out$centers) <- featnames(dfm_papers_tfidf_hamilton)
for (i in 1:k) { # loop for each cluster
cat("CLUSTER", i, "\n")
cat("Top 10 words:\n") # 10 most important terms at the centroid
print(head(sort(km_out$centers[i, ], decreasing = TRUE), n = 10))
cat("\n")
cat("Federalist Papers classified: \n") # extract essays classified
print(docnames(dfm_papers_tfidf_hamilton)[km_out$cluster == i])
cat("\n")
}## CLUSTER 1
## Top 10 words:
## armi militia revenu militari war trade taxat upon
## 6.271473 5.204072 4.634529 4.413837 4.349851 4.222969 4.065847 4.049374
## land tax
## 4.007387 3.889522
##
## Federalist Papers classified:
## [1] "No. 6" "No. 7" "No. 8" "No. 11" "No. 12" "No. 15" "No. 21" "No. 22"
## [9] "No. 24" "No. 25" "No. 26" "No. 29" "No. 30" "No. 34" "No. 35" "No. 36"
##
## CLUSTER 2
## Top 10 words:
## juri trial court crimin admiralti equiti chanceri
## 218.20102 84.74567 62.47940 42.06871 40.87463 38.24428 37.86574
## common-law probat civil
## 27.04695 27.04695 26.77843
##
## Federalist Papers classified:
## [1] "No. 83"
##
## CLUSTER 3
## Top 10 words:
## court appel jurisdict suprem juri tribun cogniz inferior
## 69.68857 35.27513 25.46591 24.79126 22.16104 21.27125 19.12214 18.76875
## appeal re-examin
## 16.21098 13.52348
##
## Federalist Papers classified:
## [1] "No. 81" "No. 82"
##
## CLUSTER 4
## Top 10 words:
## senat presid claus upon court governor offic appoint
## 5.459987 4.228173 3.530536 3.456783 3.254136 3.134332 3.071945 2.823580
## impeach nomin
## 2.737110 2.658907
##
## Federalist Papers classified:
## [1] "No. 1" "No. 9" "No. 13" "No. 16" "No. 17" "No. 23" "No. 27" "No. 28"
## [9] "No. 31" "No. 32" "No. 33" "No. 59" "No. 60" "No. 61" "No. 65" "No. 66"
## [17] "No. 67" "No. 68" "No. 69" "No. 70" "No. 71" "No. 72" "No. 73" "No. 74"
## [25] "No. 75" "No. 76" "No. 77" "No. 78" "No. 79" "No. 80" "No. 84" "No. 85"Section 5.1.4: Authorship Prediction
In a next step, we want to predict authorship for the Federalist Papers whose authorship is unknown. As the topics of the Papers differs remarkably, Imai focuses on 10 articles, prepositions and conjunctions to predict authorship.
# term frequency per 1000 words
tfm <- dfm_weight(dfm_prep, "prop") * 1000
# select words of interest
words <- c("although", "always", "commonly", "consequently",
"considerable", "enough", "there", "upon", "while", "whilst")
tfm <- dfm_select(tfm, words, valuetype = "fixed")
# average among Hamilton/Madison essays
tfm_ave <- dfm_group(dfm_subset(tfm, !is.na(author)), groups = author) /
as.numeric(table(docvars(tfm, "author")))
# bind docvars from corpus and tfm to a data frame
author_data <- data.frame(docvars(corpus_raw), convert(tfm, to = "data.frame"))
# create numeric variable that takes value 1 for Hamilton's essays,
# -1 for Madison's essays and NA for the essays with unknown authorship
author_data$author_numeric <- ifelse(author_data$author == "hamilton", 1,
ifelse(author_data$author == "madison", -1, NA))
# use only known authors for training set
author_data_known <- na.omit(author_data)
hm_fit <- lm(author_numeric ~ upon + there + consequently + whilst,
data = author_data_known)
hm_fit##
## Call:
## lm(formula = author_numeric ~ upon + there + consequently + whilst,
## data = author_data_known)
##
## Coefficients:
## (Intercept) upon there consequently whilst
## -0.2728 0.2240 0.1278 -0.5597 -0.8379## [1] 0.7175463Section 5.1.5: Cross-Validation
Finally, we assess how well the model fits the data by classifying each essay based on its fitted value.
# proportion of correctly classified essays by Hamilton
mean(hm_fitted[author_data_known$author == "hamilton"] > 0)## [1] 1
# proportion of correctly classified essays by Madison
mean(hm_fitted[author_data_known$author == "madison"] < 0)## [1] 1
n <- nrow(author_data_known)
hm_classify <- rep(NA, n) # a container vector with missing values
for (i in 1:n) {
# fit the model to the data after removing the ith observation
sub_fit <- lm(author_numeric ~ upon + there + consequently + whilst,
data = author_data_known[-i, ]) # exclude ith row
# predict the authorship for the ith observation
hm_classify[i] <- predict(sub_fit, newdata = author_data_known[i, ])
}
# proportion of correctly classified essays by Hamilton
mean(hm_classify[author_data_known$author == "hamilton"] > 0)## [1] 1
# proportion of correctly classified essays by Madison
mean(hm_classify[author_data_known$author == "madison"] < 0)## [1] 1
disputed <- c(49, 50:57, 62, 63) # 11 essays with disputed authorship
tf_disputed <- dfm_subset(tfm, is.na(author)) |>
convert(to = "data.frame")
author_data$prediction <- predict(hm_fit, newdata = author_data)
author_data$prediction # predicted values## [1] 0.73688561 -0.13805744 -1.35078839 -0.03818220 -0.27281185 0.71544514
## [7] 1.33186598 0.19419487 0.37522873 -0.20366262 0.67591728 0.99485942
## [13] 1.39358870 -1.18910700 1.20454811 0.64005758 0.91397681 -0.38337508
## [19] -0.62468302 -0.12449276 0.91560123 1.08105705 0.88535667 1.01199958
## [25] 0.08231865 0.72345360 1.08185398 0.71361766 1.47727897 1.53681013
## [31] 1.85581851 0.71928207 1.07671057 1.26171629 0.90807385 1.06348441
## [37] -0.81827318 -0.56125155 -0.27281185 -0.10304108 -0.47339830 -0.17488801
## [43] -0.60651986 -0.86943120 -1.67771035 -0.91485858 -0.13263458 -0.13495322
## [49] -0.96785069 -0.06902493 -1.46794406 -0.27281185 -0.54226168 -0.54530152
## [55] 0.04087236 -0.56414912 -1.18176926 -0.41900312 0.55206970 0.98683635
## [61] 0.59244239 -0.97797908 -0.21198475 -0.18292385 1.15647955 1.12410607
## [67] 0.78355251 0.11202677 1.12627538 0.70796660 0.34757045 0.84264937
## [73] 1.24189733 0.91619843 0.50283000 1.05488104 1.05933797 0.90593132
## [79] 0.54299626 0.64042173 0.91780877 0.31194485 0.80874960 0.73655199
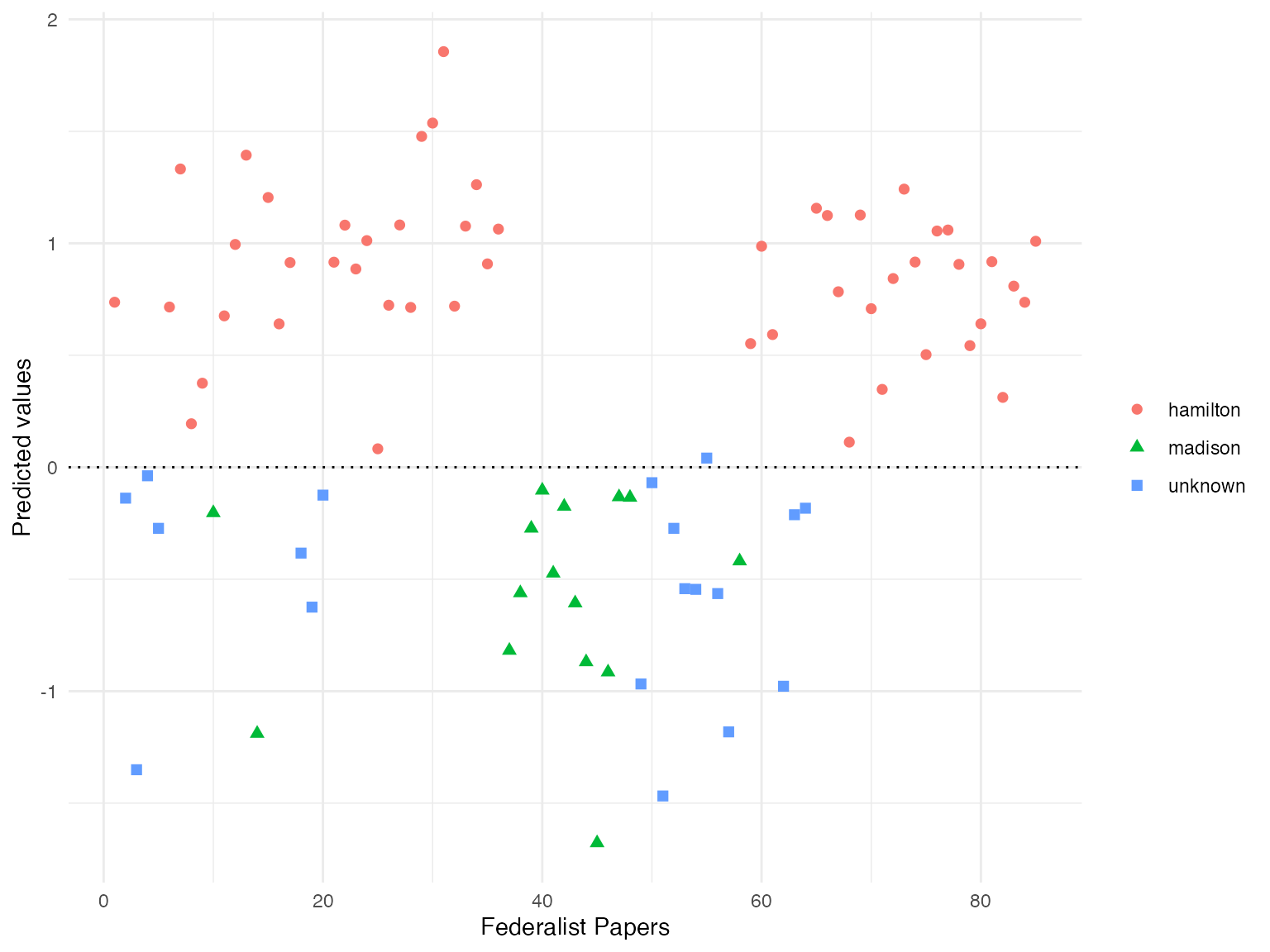
## [85] 1.00901860Finally, we plot the fitted values for each Federalist paper with the ggplot2 package.
author_data$author_plot <- ifelse(is.na(author_data$author), "unknown", as.character(author_data$author))
library(ggplot2)
ggplot(data = author_data, aes(x = paper_numeric,
y = prediction,
shape = author_plot,
colour = author_plot)) +
geom_point(size = 2) +
geom_hline(yintercept = 0, linetype = "dotted") +
labs(x = "Federalist Papers", y = "Predicted values") +
theme_minimal() +
theme(legend.title=element_blank())